LLM-Informed Gene Embeddings
Enhancing generative perturbation models with LLM-informed gene embeddings
LLM-Informed Gene Embeddings
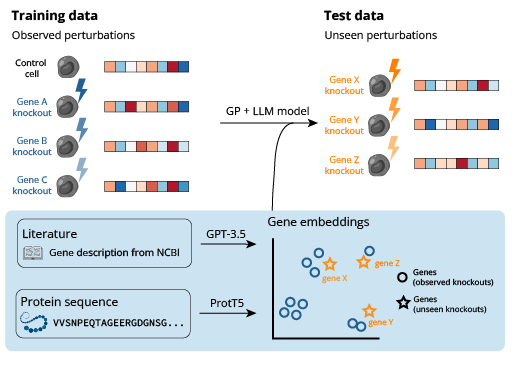
Genetic perturbations are key to understanding how genes regulate cell behavior, yet the ability to predict responses to these perturbations remains a significant challenge. While numerous generative models have been developed for perturbation data, they typically lack the capability to generalize to perturbations not encountered during training.
To alleviate this limitation, we introduce a novel methodology that incorporates prior knowledge through embeddings derived from Large Language Models (LLMs), effectively informing our predictive models with a deeper biological context. By leveraging this source of pre-existing information, our models achieve state-of-the-art performance in predicting the outcomes of single-gene perturbations.

Key Contributions
- Integration of large language model embeddings with perturbation prediction models
- State-of-the-art performance on single-gene perturbation prediction
- Improved generalization to unseen perturbations
- Novel approach to incorporating biological prior knowledge
Methods
Our approach combines transformer-based language models trained on gene sequence and function data with conditional generative models for cellular response prediction. This allows us to leverage vast amounts of pre-existing biological knowledge encoded in LLMs to inform predictions about genetic perturbations.
Results
The model demonstrates significant improvements over baseline methods, achieving higher correlation with experimental measurements and better generalization to held-out gene sets.