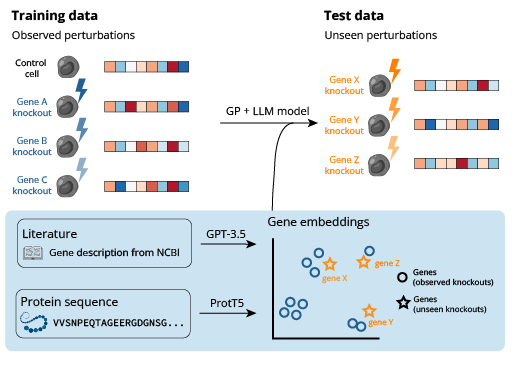
LLM-Informed Gene Embeddings
Enhancing generative perturbation models with LLM-informed gene embeddings

Enhancing generative perturbation models with LLM-informed gene embeddings
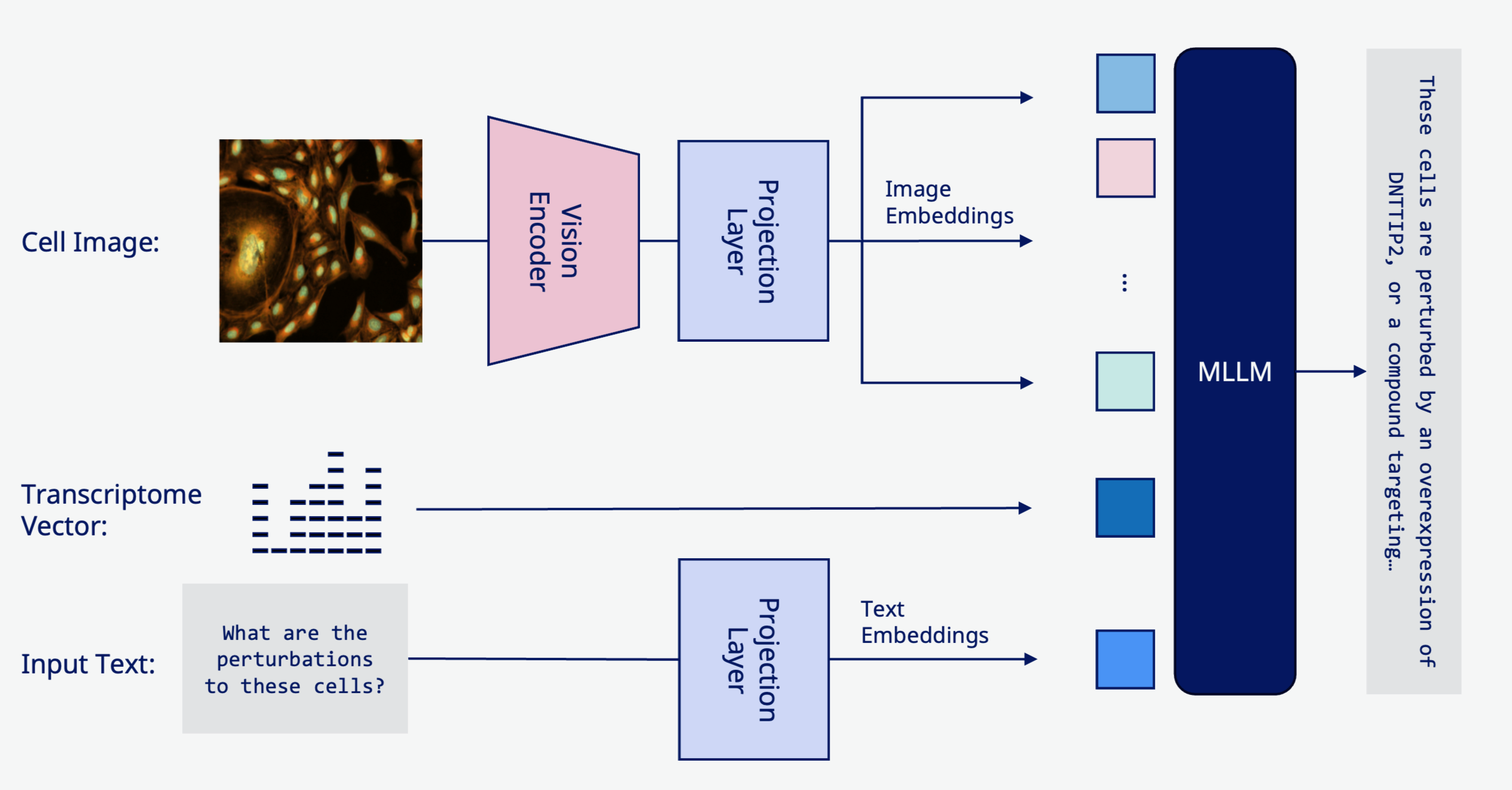
Multimodal generative models of in vitro cellular perturbations
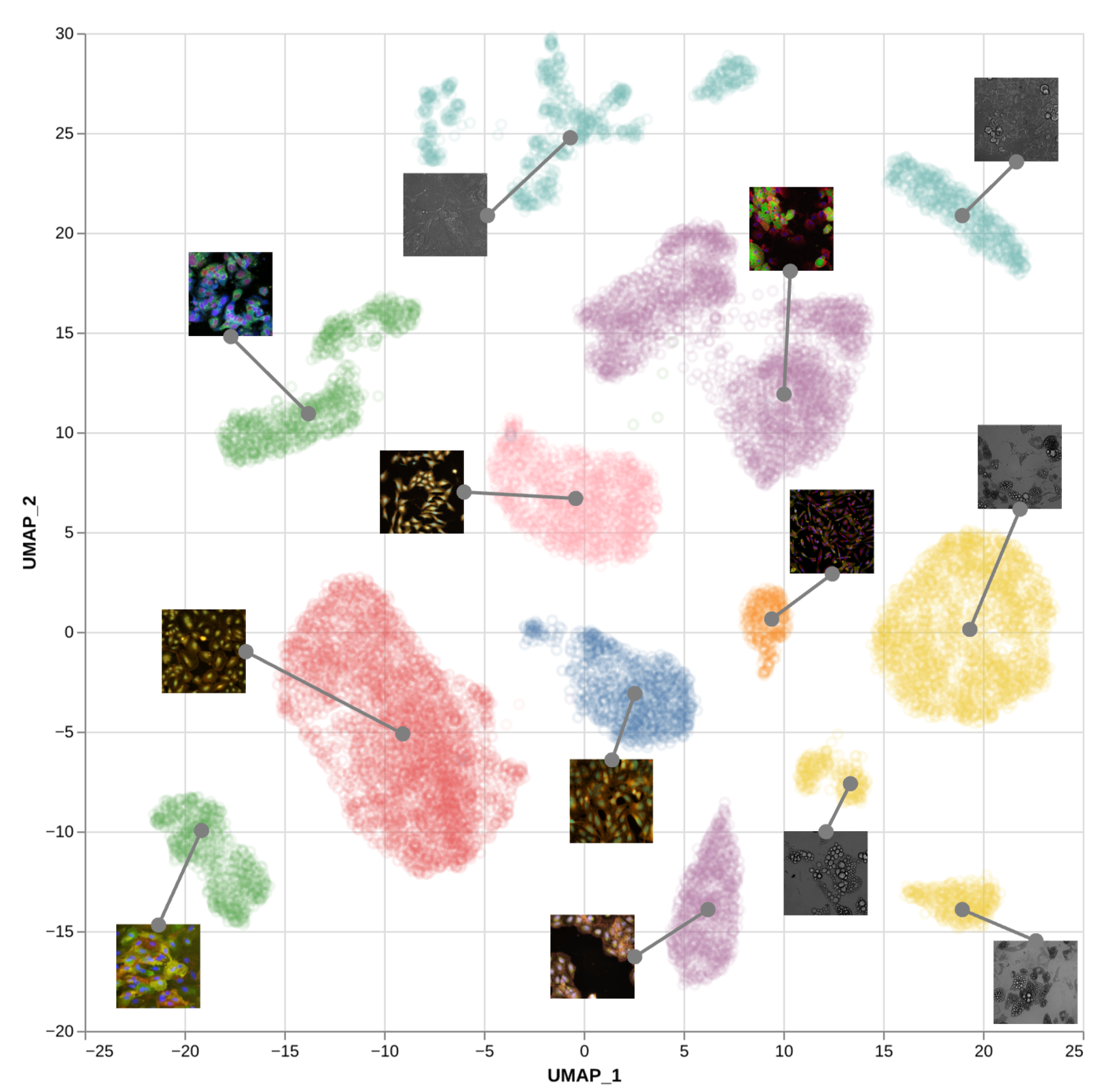
Self-supervised representation learning of cellular morphology across cell types and image modalities
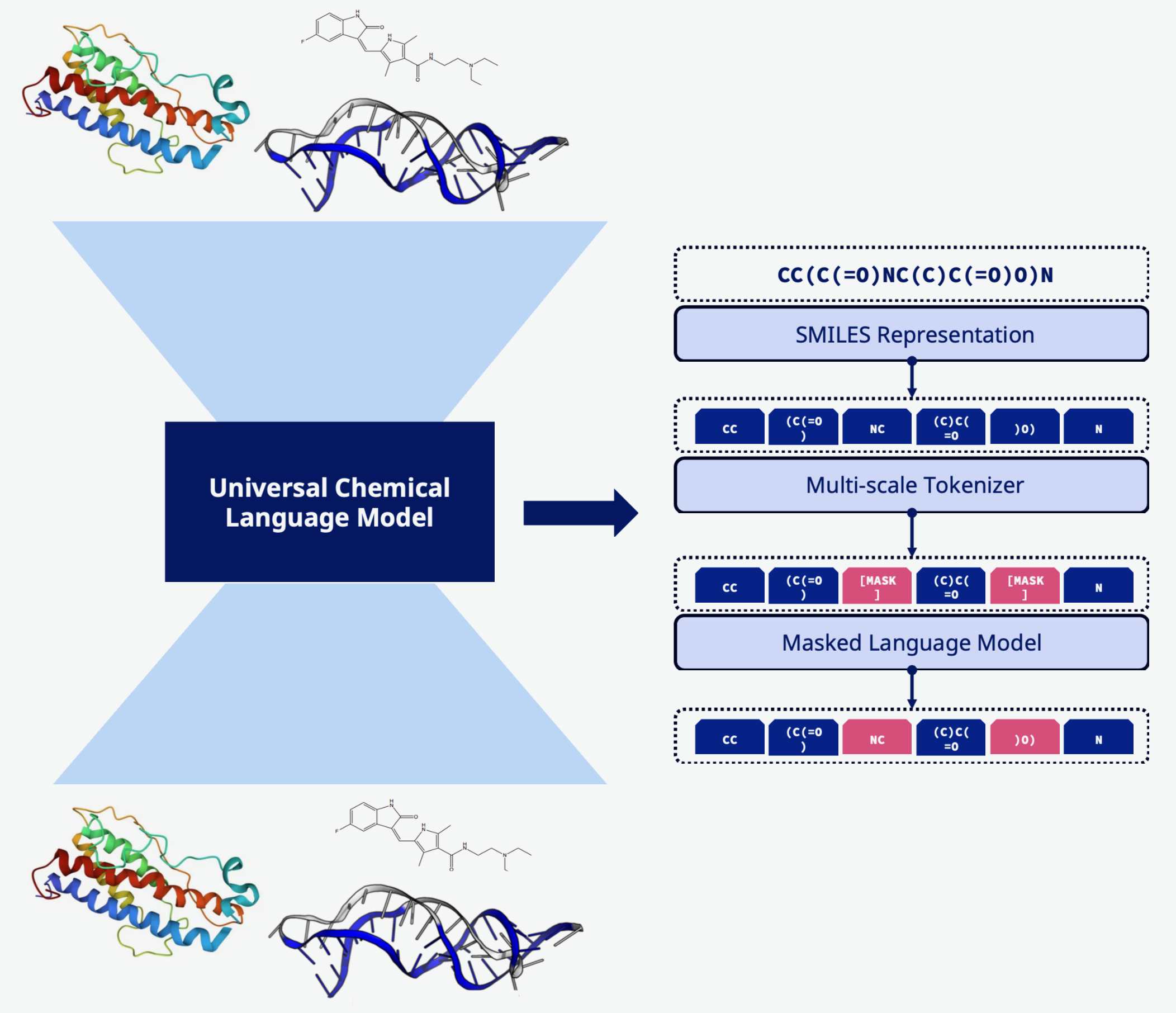
Universal language model for proteins, DNA, RNA, and small molecules
Efficient Rust-based tokenizer optimized for molecular representation
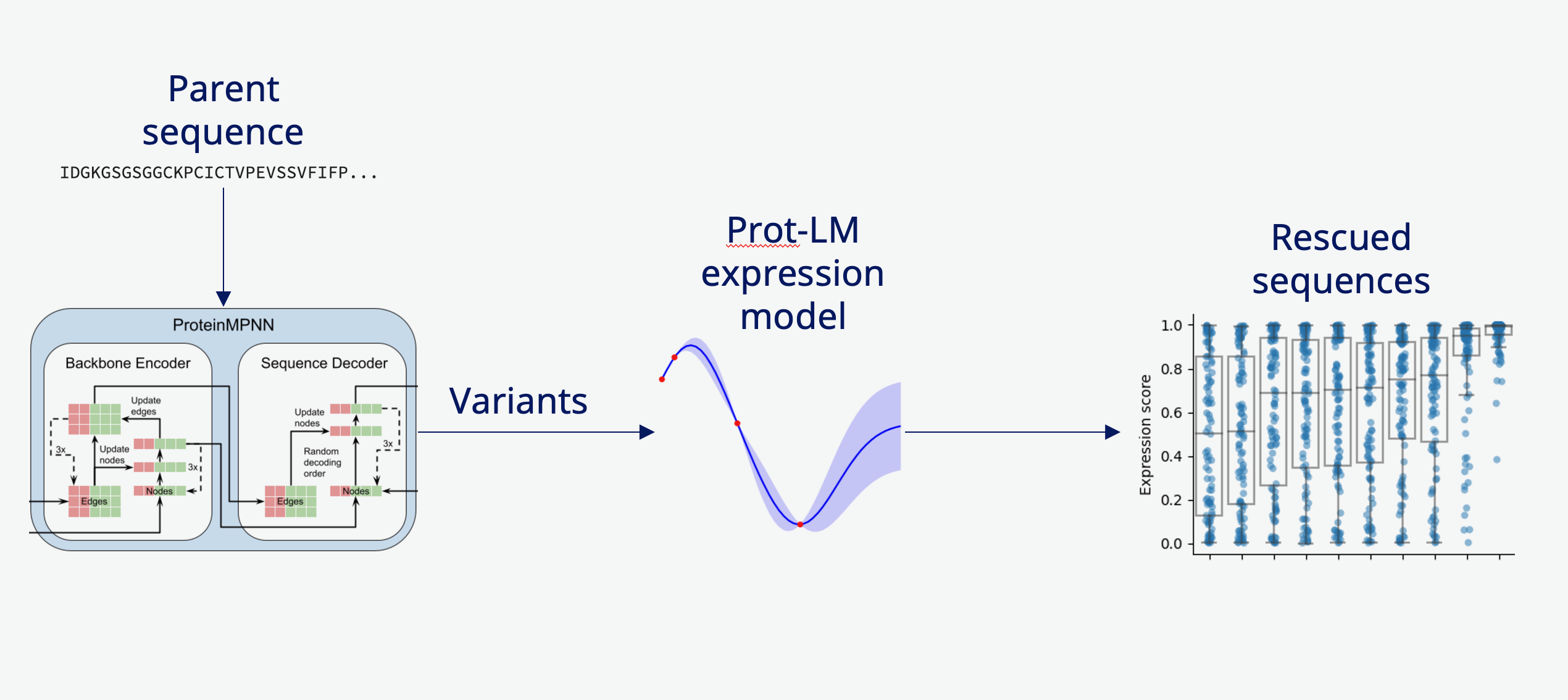
Rescuing non-expressing proteins using inverse folding and LLM-based expression prediction
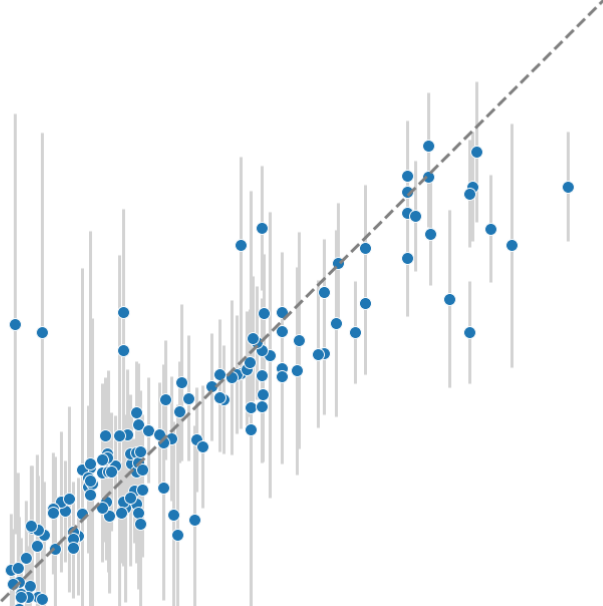
Predicting half-life and clearance rate for peptide therapeutics
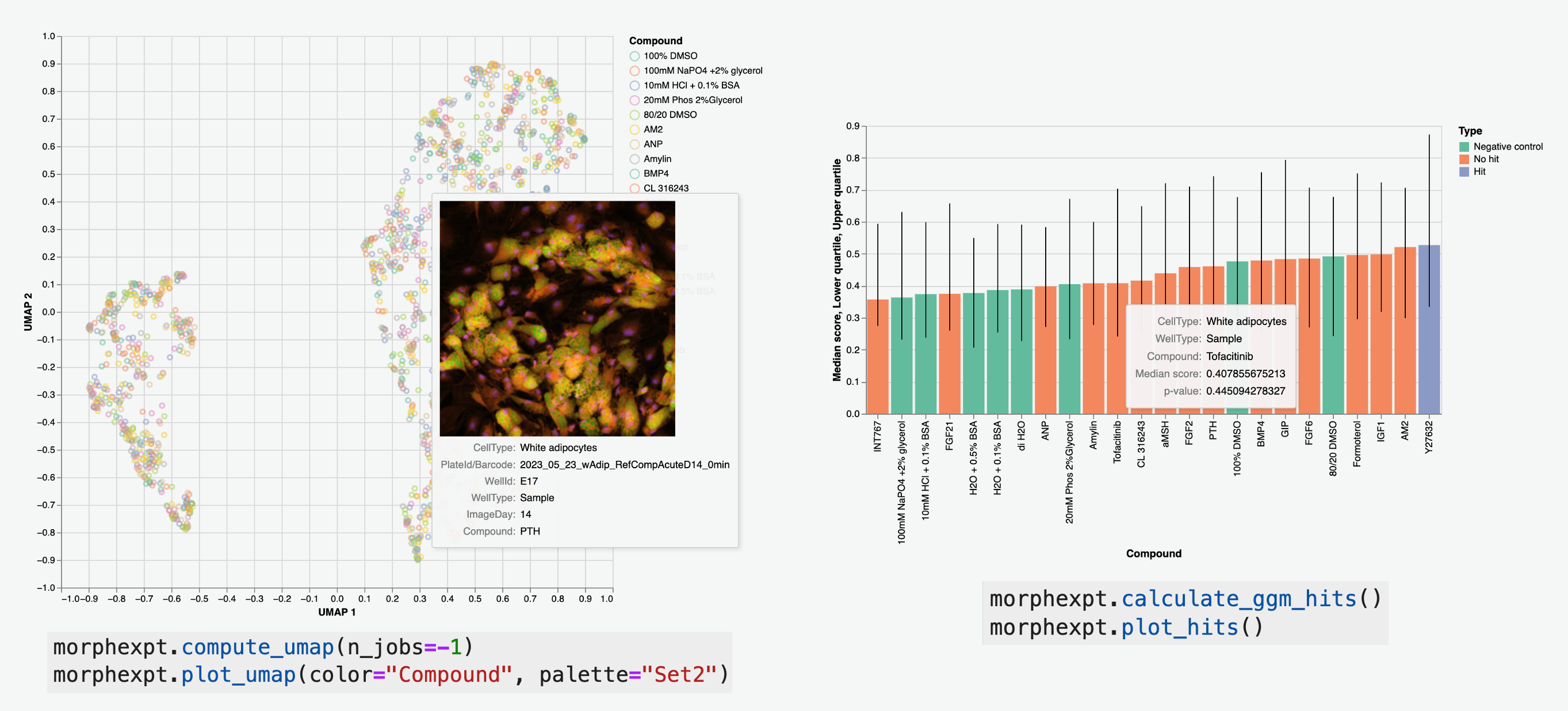
Python library for efficient microscopy data processing and analysis
