Multimodal Generative Models
Multimodal generative models of in vitro cellular perturbations
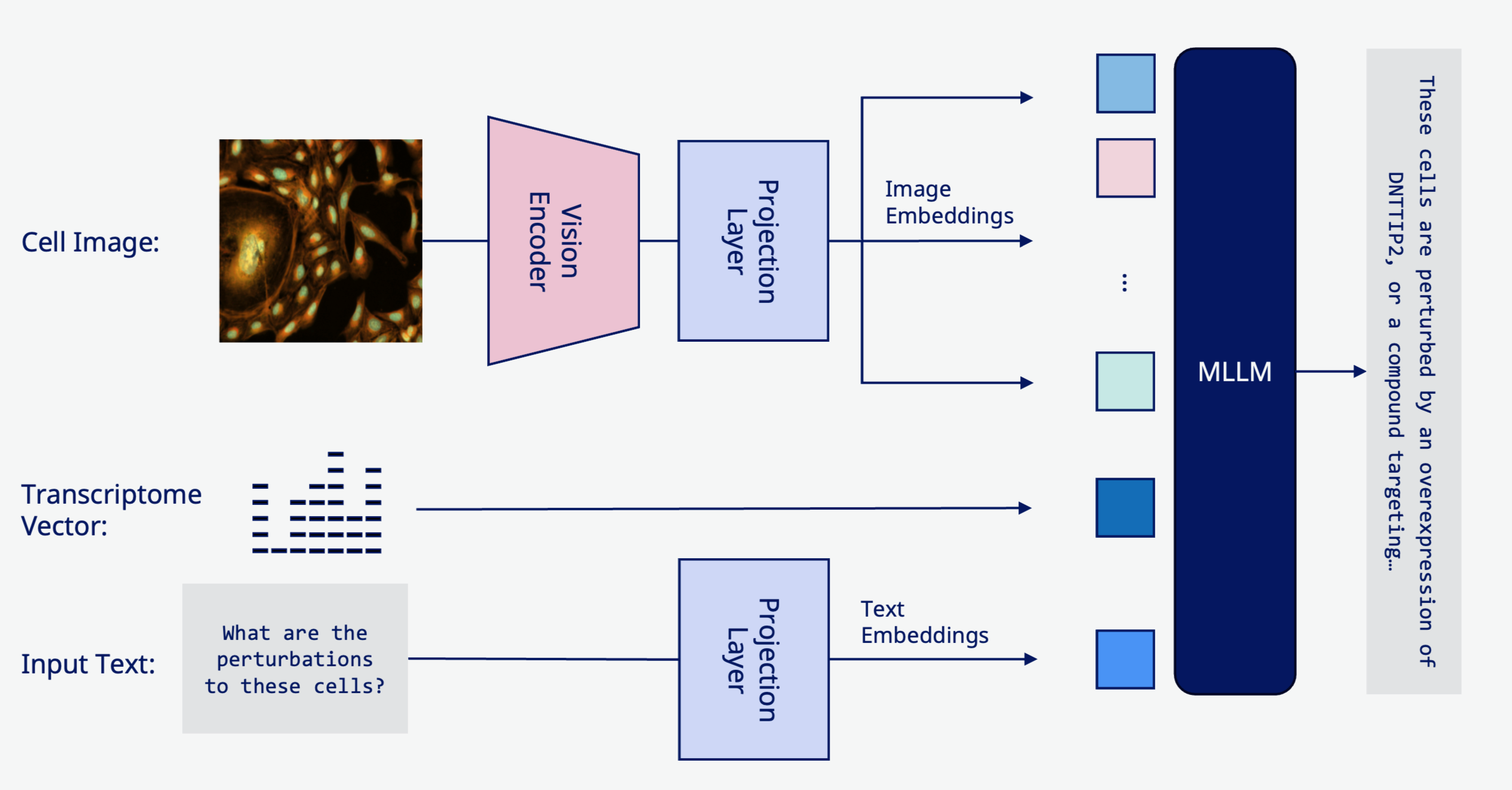
Multimodal Generative Models
Generating image-based phenotype responses to cellular perturbations in silico, and vice versa to integrate multiple screening modalities and provide in silico predictions to efficiently guide further wet-lab experiments. “Stable diffusion but for biology,” if you like.

Overview
This project develops multimodal generative models that can:
- Generate cellular image phenotypes from perturbation descriptions
- Predict perturbation conditions from observed cellular images
- Bridge multiple experimental modalities (microscopy, genomics, proteomics)
- Enable efficient in silico screening to guide wet-lab experiments
Motivation
Traditional drug discovery and biological research requires extensive wet-lab experimentation. By building generative models that can predict cellular responses across modalities, we can dramatically reduce the experimental search space and accelerate discovery.
Approach
We leverage recent advances in multimodal learning and diffusion models to build a unified framework that:
- Learns joint representations of cellular images and perturbation conditions
- Generates high-quality synthetic cellular images conditioned on perturbations
- Infers likely perturbation conditions from observed phenotypes
- Integrates data across different imaging modalities and cell types
Applications
- Drug discovery: Predict cellular responses to novel compounds
- Target identification: Identify molecular targets from phenotypic screens
- Experimental design: Optimize perturbation conditions in silico before testing
- Data integration: Combine insights from multiple screening platforms
Technical Details
The model architecture combines vision transformers for image encoding with transformer-based language models for perturbation representation, connected through a diffusion-based generative framework that enables bidirectional translation between modalities.