← Back to Projects
Universal Cell Image Embeddings
current
Self-supervised representation learning of cellular morphology across cell types and image modalities
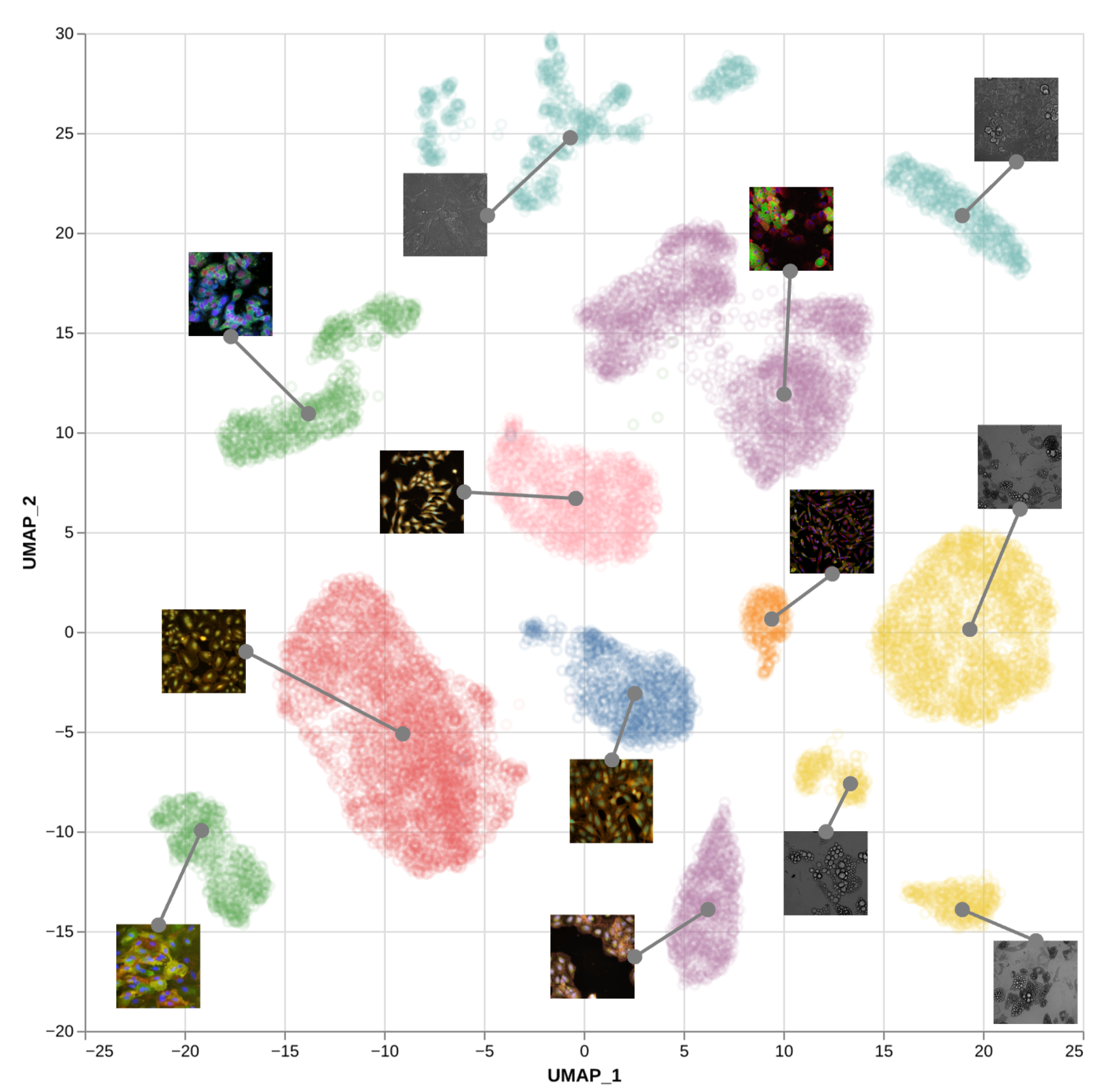
Universal Cell Image Embeddings
Self-supervised representation learning of cellular morphology / perturbation state across cell types and image modalities to build a foundational vision model capable of universally embedding all in vitro cellular image modalities.

Overview
This project aims to develop a universal foundation model for cellular imaging that can:
- Learn robust representations of cellular morphology across different cell types
- Generalize across diverse imaging modalities and experimental conditions
- Capture perturbation-induced phenotypic changes
- Enable transfer learning for downstream biological tasks
Motivation
Current cell image analysis methods often require task-specific models trained on limited datasets. A universal embedding model trained via self-supervised learning on diverse cellular imaging data can provide:
- Better generalization to new cell types and conditions
- Reduced need for labeled training data
- Consistent representations across different imaging platforms
- Foundation for multiple downstream applications
Approach
We employ self-supervised learning techniques to train vision models on large-scale cellular image datasets:
- Contrastive Learning: Learn to distinguish between different cellular states
- Multi-modal Training: Incorporate data from diverse imaging modalities
- Cross-cell-type Generalization: Train on multiple cell types to learn universal features
- Perturbation-aware Representations: Capture both baseline morphology and perturbation effects
Technical Details
- Vision transformer architectures for flexible image encoding
- Self-supervised pretraining on unlabeled cellular image datasets
- Fine-tuning strategies for specific biological applications
- Evaluation on downstream tasks including cell type classification, compound mechanism prediction, and phenotype clustering
Applications
- Drug Discovery: Rapidly screen compounds based on phenotypic similarity
- Cell Biology: Identify novel cellular states and transitions
- Quality Control: Detect anomalous cells or imaging artifacts
- Data Integration: Harmonize datasets across labs and platforms