Molecular Embeddings
Universal language model for proteins, DNA, RNA, and small molecules
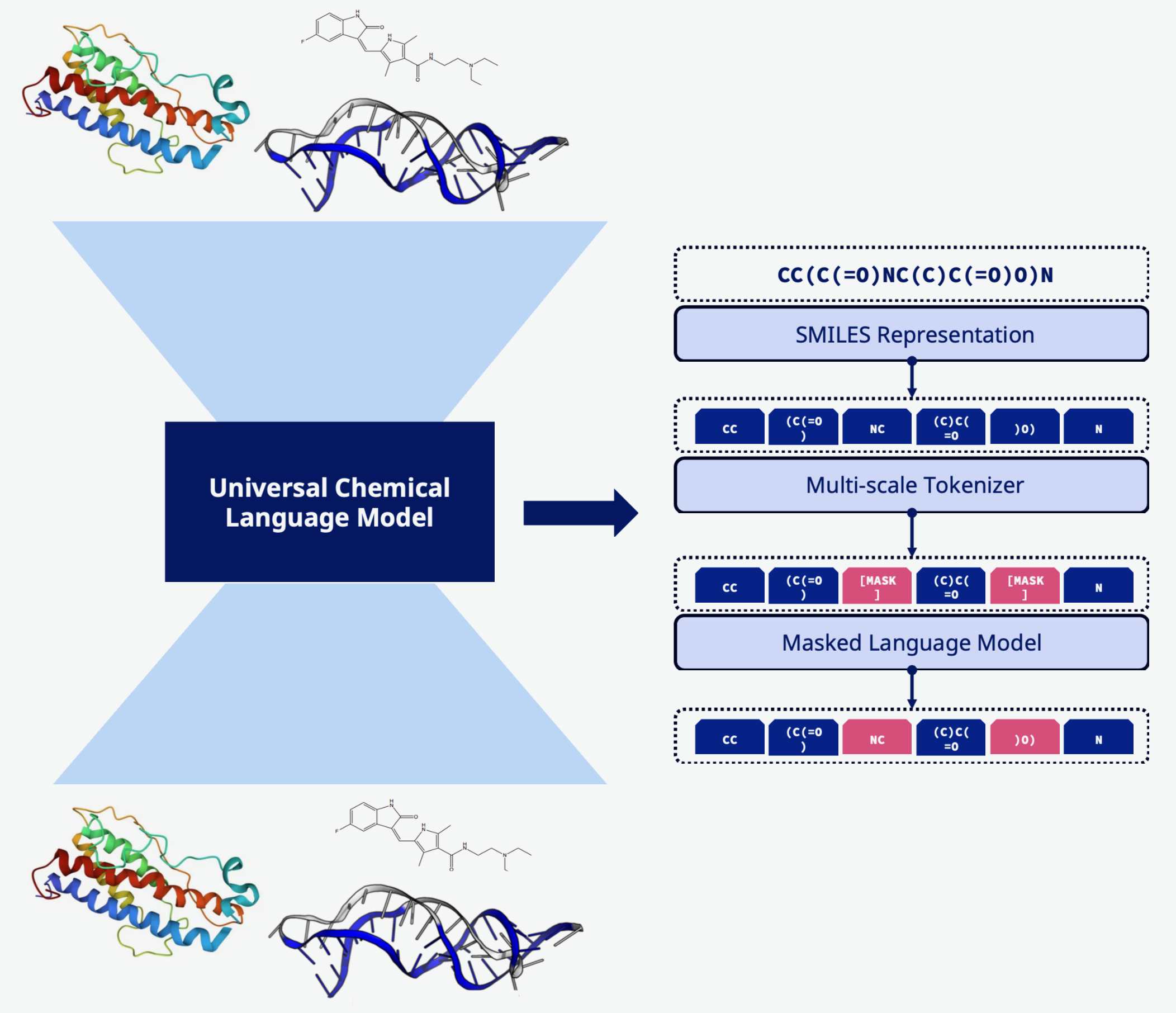
Molecular Embeddings
Protein, DNA, RNA, and small molecule language models have strong in-domain performance but lack generality: e.g., protein language models don’t understand chemical modification / non-natural amino acids, but peptides with such properties are crucial therapeutic targets. Using self-supervision and hierarchical / stochastic tokenization schemes, we train a truly general language model on the entirety of biologically-relevant molecule space, for universally applicable embeddings and generative models.

Motivation
Current molecular language models are domain-specific:
- Protein LMs trained only on natural amino acids
- Small molecule models don’t understand biological context
- RNA/DNA models are isolated from protein sequence space
- Modified residues and non-natural building blocks are poorly represented
This fragmentation limits their utility for:
- Peptide therapeutics with chemical modifications
- Protein-small molecule conjugates
- Modified nucleic acids
- Cross-domain transfer learning
Approach
We develop a unified molecular language model that:
- Universal Tokenization: Hierarchical tokenization scheme that represents proteins, nucleic acids, and small molecules in a common vocabulary
- Self-Supervised Learning: Train on vast datasets spanning all molecular domains
- Stochastic Tokenization: Multiple valid tokenizations of the same molecule for improved generalization
- Cross-Domain Transfer: Learn shared representations across molecular modalities
Technical Details
- Transformer-based architecture with specialized attention mechanisms
- Training on billions of molecular structures from multiple databases
- Novel tokenization approach that preserves chemical meaning
- Multi-task learning objectives including sequence modeling, property prediction, and structure understanding
Applications
- Drug Design: Generate novel therapeutics with desired properties
- Protein Engineering: Design proteins with non-natural modifications
- Cross-Modal Search: Find molecules with similar function across different chemical classes
- Property Prediction: Transfer learning for low-data molecular property prediction
- Therapeutic Development: Optimize peptides and small molecules simultaneously
Impact
A truly universal molecular language model enables:
- Better representation of the full chemical space relevant to biology
- Improved performance on specialized tasks through transfer learning
- Novel molecular designs that bridge traditional domain boundaries
- More efficient drug discovery pipelines