Protein Expression Rescue
Rescuing non-expressing proteins using inverse folding and LLM-based expression prediction
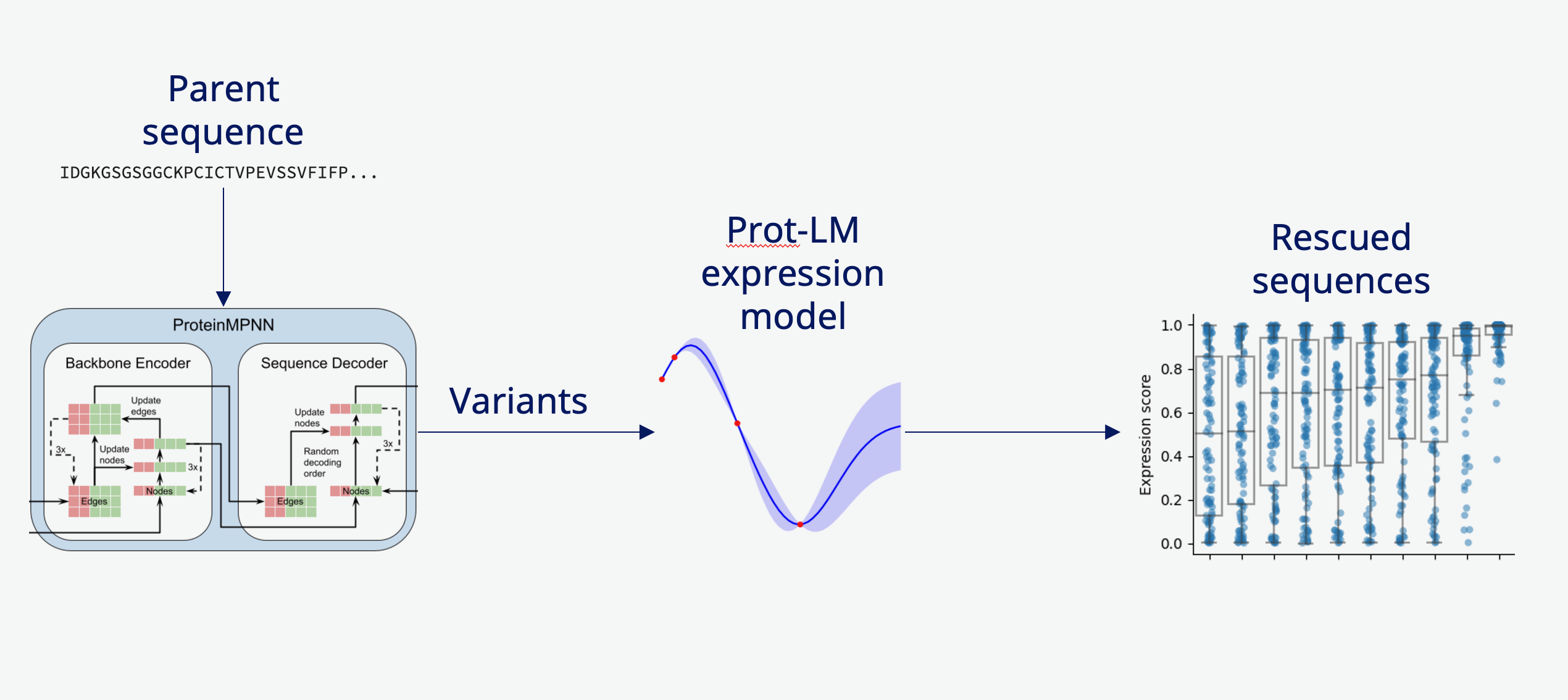
Protein Expression Rescue
Coupling inverse folding sequence generators with an in-house LLM-based expression prediction model, we rescue non- and low-expressing proteins of interest by proposing variant sequences that reliably express at 10-100x the level of the parent sequence.

Problem
A major bottleneck in protein engineering and therapeutic development is poor expression in production systems. Many proteins of interest:
- Fail to express in bacterial or mammalian hosts
- Express at levels too low for purification
- Form insoluble aggregates
- Undergo degradation
Traditional approaches require extensive trial-and-error mutagenesis, which is time-consuming and expensive.
Approach
We combine two powerful AI tools:
- Inverse Folding Models: Generate sequence variants that maintain the desired 3D structure
- Expression Prediction LLM: Predict expression levels from sequence alone
Our pipeline:
- Input: Low/non-expressing protein structure or sequence
- Generate diverse sequence variants using inverse folding (e.g., ProteinMPNN, ESM-IF)
- Score variants with our expression prediction model
- Select top candidates balancing structure preservation and high expression
- Validate experimentally
Expression Prediction Model
Our in-house LLM for expression prediction:
- Training Data: Millions of expression measurements across diverse proteins
- Architecture: Transformer-based model leveraging protein language model embeddings
- Features: Considers sequence properties, codon usage, structural features
- Output: Quantitative expression level predictions with uncertainty estimates
Results
Across multiple challenging cases:
- 10-100x expression improvement over parent sequences
- High success rate: >80% of top predictions express well
- Structure preservation: Variants maintain target fold
- Fast turnaround: Days instead of months for optimization
Applications
- Therapeutic Development: Rescue antibodies, enzymes, and novel scaffolds
- Structural Biology: Enable crystallography/cryo-EM studies requiring high protein yields
- Industrial Enzymes: Improve production economics
- Synthetic Biology: Design highly expressible parts for genetic circuits
Technical Advantages
- AI-Driven: Leverage recent advances in protein language models
- Structure-Aware: Maintain functional architecture while optimizing expression
- Uncertainty Quantification: Know confidence in predictions
- Iterative Refinement: Can be applied multiple times for further optimization
Impact
This approach dramatically reduces the time and cost of protein engineering projects by:
- Eliminating months of screening poorly expressing variants
- Reducing need for expensive mammalian expression systems
- Enabling previously intractable projects
- Accelerating therapeutic development timelines