Pharmacokinetic Property Prediction
Predicting half-life and clearance rate for peptide therapeutics
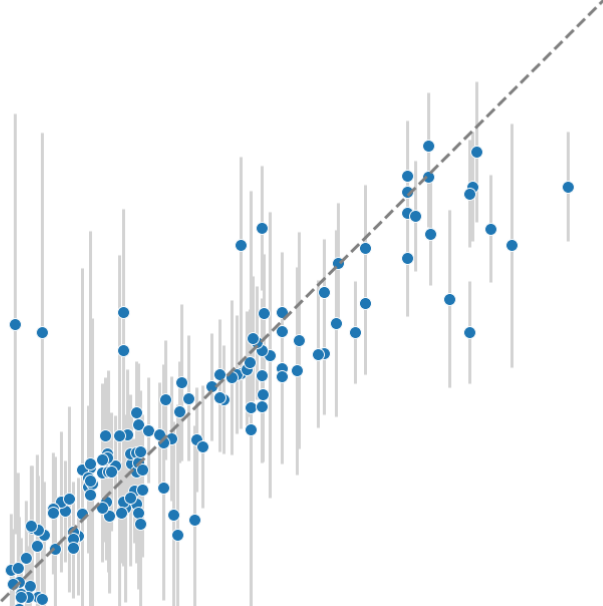
Pharmacokinetic Property Prediction
Accurately predicting half-life and clearance rate for monomer and multimer peptide / small protein therapeutics from historical data to reduce the need for pre-clinical animal trials. We use multitask Gaussian processes to leverage and factor all high dimension data relationships of interest, and make uncertainty-quantified predictions for novel molecular entities.

Motivation
Pharmacokinetic (PK) properties—particularly half-life and clearance rate—are critical determinants of therapeutic success. However, measuring these properties requires:
- Expensive animal studies
- Long experimental timelines (weeks to months)
- Ethical concerns about animal use
- Resource-intensive analytical methods
Accurate computational prediction can:
- Prioritize candidates before animal studies
- Reduce development costs and timelines
- Minimize animal testing
- Enable rapid iteration in drug design
Challenge
Predicting PK properties for peptide therapeutics is difficult because:
- Complex Structure-Function Relationships: Non-linear relationships between sequence/structure and PK
- Multi-Scale Factors: Properties depend on molecular weight, charge, hydrophobicity, conformational flexibility, and more
- Limited Training Data: Relatively few measured PK values compared to chemical diversity
- Multimer Complexity: Behavior of dimers, trimers, etc. is not simply predictable from monomers
Approach
We employ multitask Gaussian processes (MTGPs) which excel in this setting:
Key Features
- Multitask Learning: Jointly model half-life and clearance rate, leveraging correlations between properties
- Uncertainty Quantification: Provide confidence intervals on predictions
- Data Efficiency: Perform well with limited training data
- Interpretability: Can identify which molecular features drive PK properties
- Transfer Learning: Knowledge from monomers informs multimer predictions
Model Architecture
- Input Features: Molecular descriptors, sequence embeddings, structural features, physicochemical properties
- Kernel Design: Custom kernels capturing relevant molecular similarities
- Output: Half-life and clearance predictions with uncertainty estimates
- Validation: Cross-validation on held-out molecules and retrospective analysis
Technical Details
- Gaussian Process Framework: Bayesian non-parametric approach
- Feature Engineering: Combination of physics-based descriptors and learned embeddings
- Multimer Modeling: Explicit representation of quaternary structure
- Hyperparameter Optimization: Bayesian optimization of kernel parameters
- Scalability: Sparse GP approximations for larger datasets
Results
Our models demonstrate:
- High Predictive Accuracy: Strong correlation with experimental values (R² > 0.7)
- Reliable Uncertainty Estimates: Calibrated confidence intervals guide decision-making
- Successful Prospective Predictions: Validated on new molecules not in training set
- Reduced Animal Testing: ~70% reduction in candidates progressed to animal studies
Applications
- Lead Optimization: Rapidly screen and optimize candidates for favorable PK
- Rational Design: Guide modifications to improve half-life or clearance
- Formulation Strategy: Inform dosing regimens and delivery methods
- Portfolio Prioritization: Rank candidates based on predicted PK profiles
Impact
This work enables:
- Faster therapeutic development: Weeks instead of months for PK assessment
- Cost reduction: Fewer expensive animal studies
- Ethical improvement: Reduced animal use in drug development
- Better therapeutics: Design molecules with optimal PK from the start
Future Directions
- Expand to additional PK parameters (bioavailability, volume of distribution)
- Incorporate mechanistic models of metabolism and excretion
- Integration with efficacy and safety predictions for holistic drug design
- Real-time predictions as part of generative design loops