High Throughput Image Analysis Library
Python library for efficient microscopy data processing and analysis
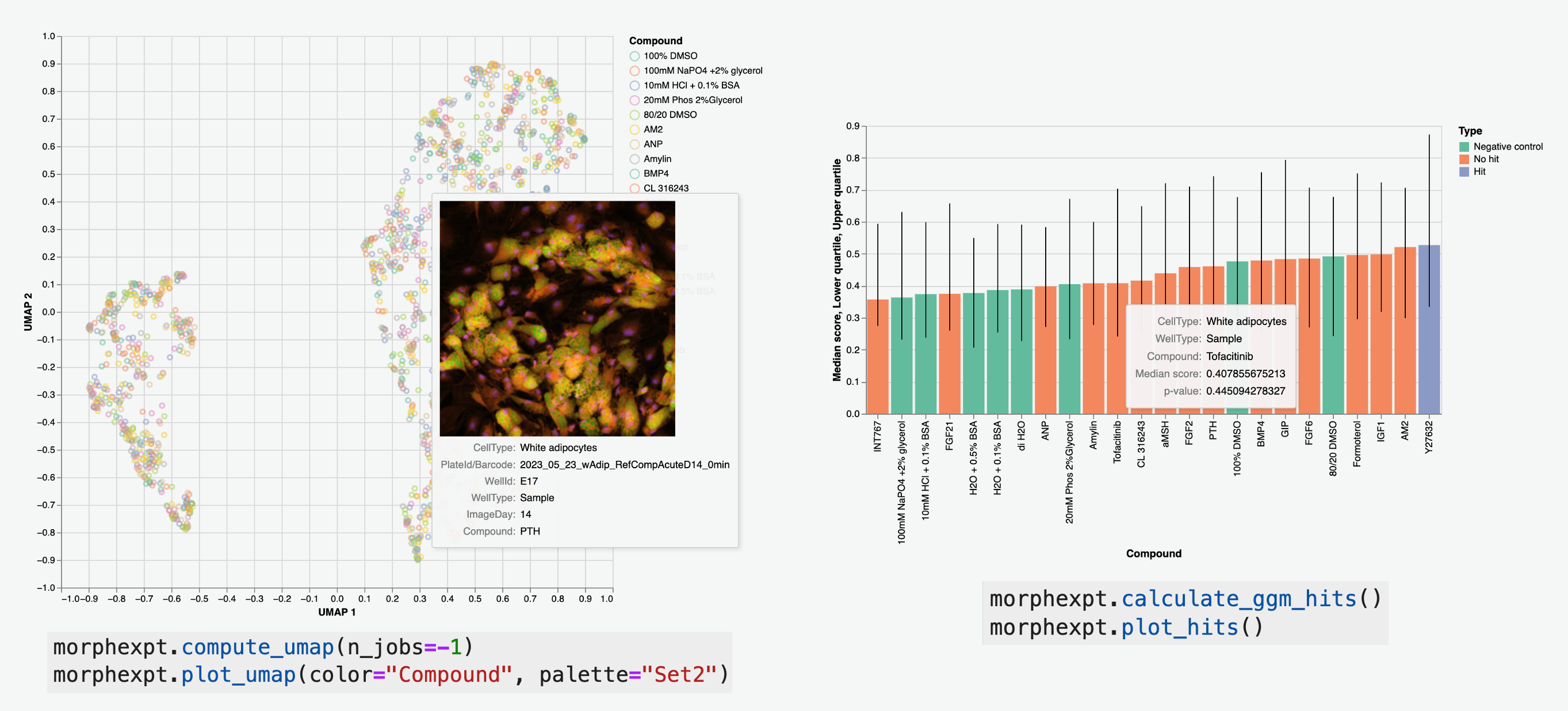
High Throughput Image Analysis Library
Foundational image / multimodal models are data hungry; to efficiently feed them with new data it’s imperative to make data QC, collation, processing, analysis, and modeling as modular, performant, and painless as possible. A Python library with templated HPC workflows, a modular CLI, pre-prepared analysis notebooks, and import/export image QC enables rapid, reproducible, and verifiable microscope → knowledge distillation, as well as efficient data curation for training large foundational models.

Motivation
Modern microscopy generates massive datasets:
- Terabytes of images per experiment
- Thousands of experimental conditions
- Multiple imaging modalities and timepoints
- Diverse cell types and perturbations
Processing this data requires:
- Quality Control: Automated detection of imaging artifacts and errors
- Standardization: Consistent preprocessing across experiments
- Scalability: HPC-enabled workflows for large datasets
- Reproducibility: Tracked, versioned, documented pipelines
- Accessibility: Easy-to-use interfaces for diverse users
Solution
A comprehensive Python library providing:
Core Components
-
Modular CLI: Command-line interface for common operations
- Image ingestion and format conversion
- Batch preprocessing and normalization
- Feature extraction and analysis
- Model training and inference
-
HPC Workflow Templates: Pre-configured for common cluster systems
- SLURM, PBS, SGE job submission
- Distributed processing with Dask
- Containerized workflows (Docker, Singularity)
- Automatic resource allocation
-
Image QC Pipeline: Automated quality assessment
- Focus detection
- Illumination correction validation
- Artifact detection (debris, bubbles, edge effects)
- Statistical outlier identification
- Interactive QC dashboards
-
Analysis Notebooks: Jupyter notebooks for common analyses
- Exploratory data analysis
- Phenotype clustering and visualization
- Statistical comparisons
- Model evaluation and interpretation
-
Data Curation Tools: Prepare datasets for ML training
- Balanced sampling strategies
- Train/val/test splitting
- Metadata management
- Export to standard formats (HDF5, Zarr, TFRecord)
Key Features
Performance
- Parallel Processing: Multi-threaded image operations
- Memory Efficiency: Streaming processing for large images
- GPU Acceleration: CUDA-enabled preprocessing when available
- Lazy Evaluation: Process only what’s needed
Reproducibility
- Configuration Management: YAML-based experiment configs
- Provenance Tracking: Automatic logging of all operations
- Version Control Integration: Git-friendly outputs
- Containerization: Reproducible environments
Extensibility
- Plugin Architecture: Easy to add custom operations
- Modular Design: Use only the components you need
- API-First: Programmatic access to all functionality
- Integration-Ready: Works with scikit-image, napari, cellpose, etc.
Architecture
morphexpt/
├── cli/ # Command-line interface
├── io/ # Data import/export
├── preprocess/ # Image preprocessing
├── qc/ # Quality control
├── features/ # Feature extraction
├── workflows/ # HPC templates
├── notebooks/ # Analysis templates
└── utils/ # Shared utilitiesWorkflow Example
# Ingest raw microscopy data
morphexpt ingest --source /path/to/images --output data/raw
# Run QC pipeline
morphexpt qc --input data/raw --output data/qc_report
# Preprocess images
morphexpt preprocess --input data/raw --output data/processed \
--operations normalize,background_subtract,register
# Extract features
morphexpt features --input data/processed --output data/features \
--methods morphology,texture,intensity
# Prepare ML training dataset
morphexpt curate --input data/processed --output data/train \
--split 0.7/0.15/0.15 --format zarrApplications
Foundation Model Training
- Large-Scale Curation: Process millions of images for pre-training
- Quality Filtering: Ensure clean training data
- Efficient Storage: Optimized formats for fast loading
- Metadata Tracking: Maintain experimental context
High-Content Screening
- Batch Analysis: Process entire screening campaigns
- Hit Identification: Statistical analysis of phenotypes
- Dose-Response: Automated curve fitting
- Visualization: Generate publication-ready figures
Collaborative Research
- Standardization: Common processing across labs
- Sharing: Package datasets with processing code
- Benchmarking: Compare methods on common data
- Teaching: Notebooks for education and training
Impact
This library enables:
- 10-100x speedup in microscopy data processing
- Reduced errors through automated QC
- Better reproducibility in computational imaging
- Faster iteration on ML models
- Democratized access to advanced image analysis
Technical Stack
- Core: Python 3.9+, NumPy, scikit-image
- Distributed: Dask, Ray
- I/O: Zarr, HDF5, OME-TIFF
- Visualization: Matplotlib, napari
- CLI: Click, Rich
- Testing: Pytest, hypothesis
Future Development
- Integration with more ML frameworks (PyTorch Lightning, JAX)
- Real-time analysis during acquisition
- Cloud-native processing (AWS, GCP)
- Active learning for data curation
- Automated hyperparameter optimization